MD plot showing the log-fold change and average abundance of each gene.... | Download Scientific Diagram
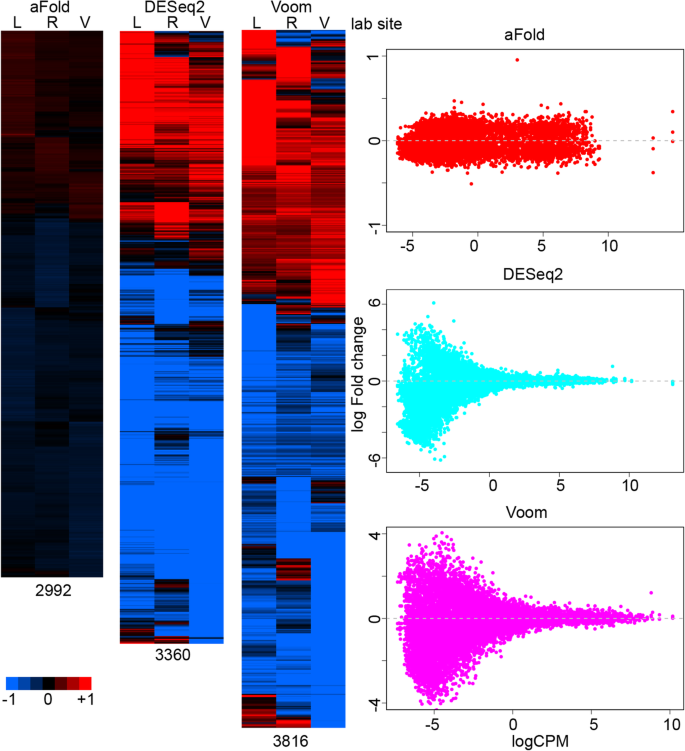
aFold – using polynomial uncertainty modelling for differential gene expression estimation from RNA sequencing data | BMC Genomics | Full Text

Advantages of RNA‐seq compared to RNA microarrays for transcriptome profiling of anterior cruciate ligament tears - Rai - 2018 - Journal of Orthopaedic Research - Wiley Online Library

Differentially expressed protein groups (q < 0.4 and Log 2 fold change... | Download Scientific Diagram

MD plot showing the log-fold change and average abundance of each gene.... | Download Scientific Diagram
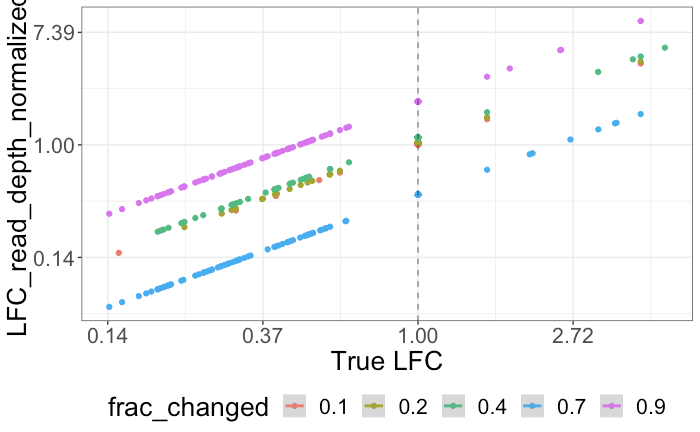
Relative vs Absolute: How to Do Compositional Data Analyses. Part — 2 | by Krishna Yerramsetty | Towards Data Science

Differentially expressed protein groups (q < 0.4 and Log 2 fold change... | Download Scientific Diagram

Differentially expressed protein groups (q < 0.4 and Log 2 fold change... | Download Scientific Diagram

Transite: A Computational Motif-Based Analysis Platform That Identifies RNA-Binding Proteins Modulating Changes in Gene Expression - ScienceDirect
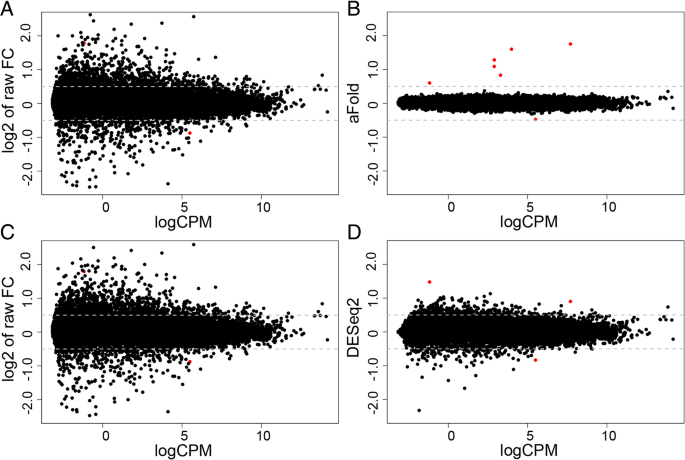
aFold – using polynomial uncertainty modelling for differential gene expression estimation from RNA sequencing data | BMC Genomics | Full Text